Discovery of Enzymes to Catalyze Pericyclic Reactions.
A couple of days ago I attended a lecture by K.N. Houk which inspired me to look into a subject about which I have not thought for a long time, my days in synthetic chemistry being far behind me; somehow I drifted elsewhere. As a result, partly out of nostalgia, partly out of curiosity, a partly because of my disappointed realization that I could have been, should have been, more, I was inspired to check out this paper pointing to a topic about which I'd only mused casually, this many years ago: Masao Ohashi, Fang Liu, Yang Hai, Mengbin Chen, Man-cheng Tang, Zhongyue Yang, Michio Sato, Kenji Watanabe, K. N. Houk & Yi Tang, SAM-dependent enzyme-catalysed pericyclic reactions in natural product biosynthesis. Nature 549, 502–506 (2017).
The question is that while pericyclic reactions - reactions that feature, in the thermal case, 2n + 2 electrons moving in a quasi aromatic transition state (i.e., 2, 6, 10, 12...electrons, most commonly 6) in such a way that the stereochemistry is tightly controlled - are frequently used in laboratory syntheses of complex natural products, in nature, these types of reactions are not particularly well characterized in biological systems. I sort of mused about this in a half serious way; it never occurred to me to seriously look into the matter. In fact, I never even thought as to whether biological pericyclic reactions were common or even known.
It turns out they are known.
The abstract of the paper points, quite well, to the question I never more than casually asked myself:
Pericyclic reactions—which proceed in a concerted fashion through a cyclic transition state—are among the most powerful synthetic transformations used to make multiple regioselective and stereoselective carbon–carbon bonds1. They have been widely applied to the synthesis of biologically active complex natural products containing contiguous stereogenic carbon centres2,3,4,5,6. Despite the prominence of pericyclic reactions in total synthesis, only three naturally existing enzymatic examples (the intramolecular Diels–Alder reaction7, and the Cope8 and the Claisen rearrangements9) have been characterized. Here we report a versatile S-adenosyl-L-methionine (SAM)-dependent enzyme, LepI, that can catalyse stereoselective dehydration followed by three pericyclic transformations: intramolecular Diels–Alder and hetero-Diels–Alder reactions via a single ambimodal transition state, and a retro-Claisen rearrangement. Together, these transformations lead to the formation of the dihydropyran core of the fungal natural product, leporin10.
From the paper's introduction:
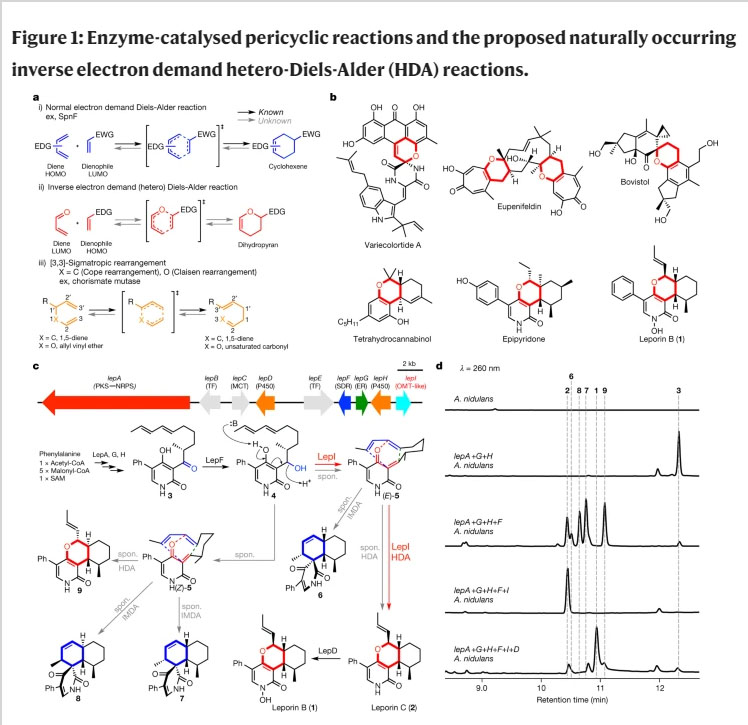
Naturally existing enzymatic pericyclic reactions are rare12,13,14. Indeed, only a handful of enzymes that can catalyse these reactions have been characterized over the past five decades (Fig. 1a)7,8,9,12,13,14, even though pericyclic reactions have been proposed as key transformations in the biosynthesis of many polycyclic natural products12,15,16. We sought an enzyme-catalysed inverse electron demand hetero-Diels–Alder (HDA) reaction17 that constructs heterocycles in natural products (Fig. 1a). The HDA reaction has been proposed as a key biotransformation yielding dihydropyran cores, which are prevalent structural features in natural products that include the cytotoxic leporin B (1) from Aspergillus species (Fig. 1b)10,15,16,18. The biomimetic synthesis of the dihydropyran core in leporin uses the E/Z geometric mixture of the unstable ?-quinone methide19 intermediate 5 generated from the dehydration of alcohol 4. The uncatalysed process gives a mixture of the minor desired HDA adduct leporin C (2) and major other regio- and stereoisomeric intramolecular Diels–Alder (IMDA) and HDA adducts (Fig. 1c)20. It was therefore proposed that an enzyme must be encoded in the biosynthetic pathway of leporins to catalyse the HDA cycloaddition in a stereoselective fashion and to suppress the IMDA reaction to afford the dihydropyran core in 2 (Fig. 1c)14.
A figure:
The caption:
a, Examples (ex) of pericyclic reactions and enzymes that can catalyse pericyclic reactions. Known and unknown enzymatic reactions are shown in black and grey arrows, respectively. EDG, electron donating group; EWG, electron withdrawing group; HOMO, highest occupied molecular orbital; LUMO, lowest unoccupied molecular orbital; b, The structures of natural products containing dihydropyran, which would be biosynthesized by HDA reaction. Variecolortide A is naturally racemic; the relative stereochemistry of epipyridone and leporin B are shown. c, The putative leporin biosynthetic gene cluster in A. flavus and the assignment of encoded genes and biosynthetic pathway of leporins. PKS–NRPS, polyketide synthase–non-ribosomal peptide synthetase; TF, transcription factor; MCT, monocarboxylate transporter; SDR, short-chain dehydrogenase/reductase; ER, enoylreductase; OMT, O-methyltransferase. The structures show the relative stereochemistry. d, Analysis of metabolites from the transformants of A. nidulans by HPLC. The peak at 12?min corresponds to the tetramic acid product that is biosynthesized by LepA (PKS–NRPS) and LepG (ER).
Some more text:
The biosynthetic gene cluster of leporin B (1) in Aspergillus flavus has been reported and genetically verified (Fig. 1c)10. However, no clear enzyme candidate that can catalyse the pericyclic reaction was apparent in the cluster. To identify the enzyme responsible for this biotransformation, we heterologously reconstituted the leporin B (1) biosynthetic pathway in Aspergillus nidulans (Fig. 1c, d)21. As shown in Fig. 1d, coexpression of the polyketide synthase?non-ribosomal peptide synthetase (PKS?NRPS) LepA, the partnering enoyl reductase (ER) LepG, and the ring-expansion P45022 LepH led to the biosynthesis of the ketone 3. Additional coexpression with the short chain dehydrogenase/reductase (SDR) LepF, which is hypothesized to reduce 3 to the alcohol 4, led to a mixture of HDA products including the desired dihydropyran 2 as a minor product and the diastereomer 9, as well as the spirocyclic IMDA products 6–8 (Fig. 1c, d). Among these products, 2 and 6 are proposed to be derived from the quinone methide (E)-5, while 7–9 are from (Z)-5 (Fig. 1c). These results are consistent with biomimetic synthetic observations, and indicate that in order to biosynthesize 2 as the desired pericyclic reaction product, enzymatic stereocontrol of dehydration of 4 to (E)-5, as well as control of the subsequent pericyclic reaction, are required.
The only remaining annotated enzyme in the gene cluster is LepI, which is predicted to be an O-methyltransferase (OMT) with a well-conserved SAM binding site even though no O-methylation step is required for leporin B (1) biosynthesis. When lepI was introduced into the A. nidulans strain that produced the various pericyclic products derived from reduction of 3, we were surprised to observe the exclusive production of 2 without any other products (Fig. 1d). Further addition of the P450 lepD yielded the final product 1, thereby completing heterologous pathway reconstitution (Fig. 1c, d). To first verify the function of SDR LepF, recombinant protein was expressed from Saccharomyces cerevisiae and assayed in the presence of 3 and NADPH, which yielded a single product 4 corresponding to the reduced compound (Extended Data Fig. 1). To obtain sufficient 4 for assay with LepI, we reduced 3 with NaBH4 which gave both 4 and diastereomer 4? in a ratio of approximately 1:1 (Extended Data Figs 1, 2). Each isomer was isolated and immediately added to LepI expressed and purified from Escherichia coli. Both 4 and 4? dehydrated spontaneously in the absence of LepI and afforded a mixture of IMDA (6–8) and HDA (2 and 9) products, with 2 being a very minor product (Fig. 2a). However, when LepI was added to 4, complete conversion to 2 was accomplished in the absence of any added cofactors (Fig. 2a). In contrast, addition of LepI to 4? had only a small effect on product profile. The collective in vivo and in vitro data therefore point to LepI being solely responsible for formation of 2 starting from 4, which requires stereoselective dehydration to yield (E)-5 and subsequent HDA reaction to 2...
"SAM" is a nucleoside, adenosine, that has alkylated a methionine, "S-adenosyl methionine," a sulfonium ion. It is a moiety that is often found in biosynthetic methylations. Methionine itself is one of the 20 coded amino acids.
This paper is highly cited. A description of the mechanism and the nature of the SAM cofactor's role in the mechanism can be found here: Min Chang, Yu Zhou, Hao Wang, Zihe Liu, Yi Zhang, Yue Feng,
Crystal structure of the multifunctional SAM-dependent enzyme LepI provides insights into its catalytic mechanism,
Biochemical and Biophysical Research Communications, Volume 515, Issue 2, 2019, Pages 255-260, and probably elsewhere.
This is a little esoteric, but it made me think a lot outside of the little box in which I live and to reflect a bit on my life and despite much happiness, some regrets.
